Background: Subtle, prognostically-meaningful ECG features may not be apparent to physicians. In the course of supervised machine learning (ML) training, many thousands of ECG features are identified. These are not limited to conventional ECG parameters and morphology. These novel NN-derived ECG features may have clinical, phenotypic and genotypic associations and prognostic significance.
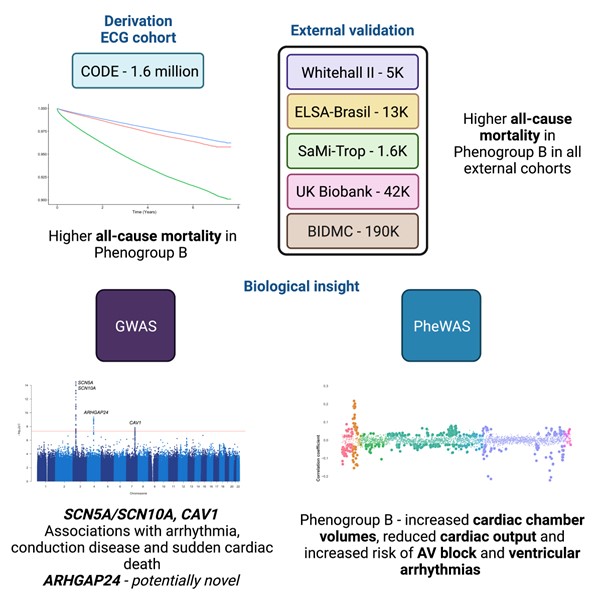
Methods and results: We extracted 5,120 NN-derived ECG features from an AI-ECG model trained for six simple diagnoses and applied unsupervised machine learning to identify three phenogroups. The derivation set, the Clinical Outcomes in Digital Electrocardiography (CODE) cohort (n=1,558,421), is a database of ECGs recorded in primary care in Brazil. There were five external validation datasets; Whitehall II (civil servants, n=5,066), UK Biobank (volunteers, n=42,386), ELSA-Brasil (public servants, n=13,739). SaMi-Trop (patients with chronic Chagas
cardiomyopathy, n=1,631) and Beth Israel (BIDMC) (secondary care population, n=188,972). In the derivation cohort (CODE), the three phenogroups had significantly different mortality profiles. After adjusting for known covariates, phenogroup B had a 1.2-fold increase in long-term mortality compared to phenogroup A (HR: 1.20, 95% CI 1.17–1.23, p < 0.0001). Importantly, the predictive ability of the phenogroups was retained in a group without any of the six diagnoses for which CODE-CNN was originally trained and in a group with clinician-reported normal ECGs. We then externally validated our findings in five diverse, multi-ethnic cohorts. We found phenogroup B had a significantly greater risk of mortality in all five external cohorts. We performed a phenome-wide association study (PheWAS) of clinical diagnoses in the BIDMC dataset. Phenogroup B was associated with a significantly higher rate of future atrial fibrillation, ischaemic heart disease, atrioventricular block, cardiomyopathy, ventricular tachycardia and cardiac arrest. PheWAS of imaging phenotypes showed phenogroup B was associated with increased cardiac chamber volumes and carotid intima-media thickness, and decreased cardiac output and left ventricular strain. A single-trait genome-wide association study (GWAS) was conducted. The GWAS yielded four loci. SCN10A, SCN5A and CAV1 have well described roles in cardiac conduction and arrhythmia. The cardiac role of ARHGAP24 is unclear and is a potentially novel finding. In order to better understand the reasons for phenogroup classification by the hybrid ML model, we used a modified Grad-CAM approach. The terminal part of the QRS complex and T wave were most important for identification of the high-risk phenogroup (phenogroup B). These may represent myocardial conduction slowing and repolarization heterogeneity respectively.
Conclusion: We describe the use of NN-derived ECG features, to identify prognostically-significant phenogroups from the 12-lead ECG. We explored the biological basis underlying the difference in prognosis between the phenogroups, and identified phenotypic and genotypic associations through PheWAS and GWAS. We validated our findings in five external datasets across two continents and diverse patient populations. NN-derived ECG features have important applications beyond the original model from which they are derived and may be transferable and applicable for risk prediction in a wide range of settings, in addition to mortality prediction. ❑
Figure 1