Background: Adoption of electronic health records (EHR) improved the availability of medical documentation for research purposes. However, significant proportion of data is in textual information that cannot be utilized for scientific purposes until it is analyzed through manual chart review. Utilization of only structured data from EHR is insufficient for comprehensive cohort characterization and of variable quality. Natural language processing can be utilized to unlock valuable data from textual format.
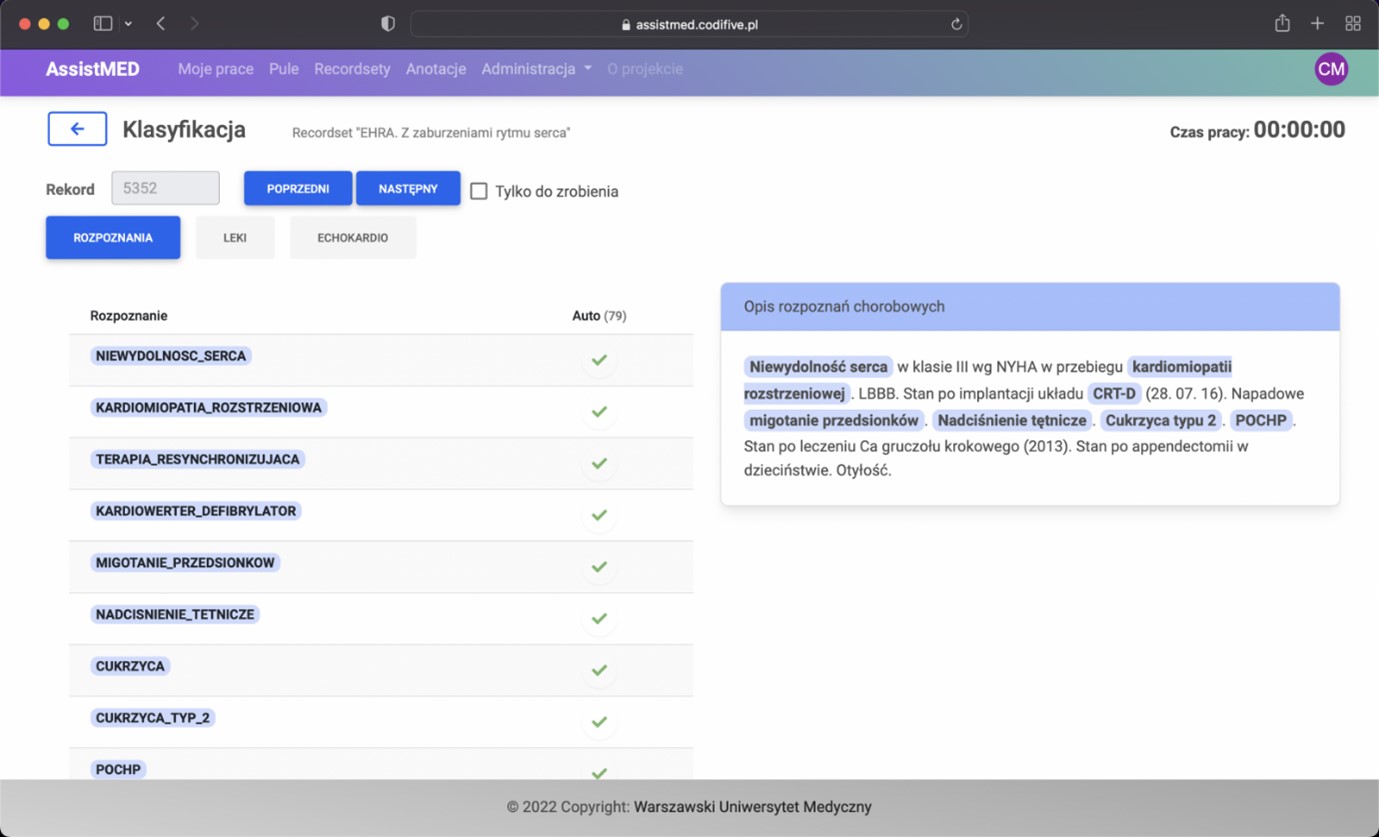
Purpose: We developed a comprehensive text-processing tool for cardiology field. The algorithm employs advanced text processing based on a specifically designed, vast database of medical terminology, drug lists and echocardiography parameters with data structure tailored to the needs of clinical researchers. The algorithm can automatically analyze 3 types of textual data which are universal parts of discharge summary in Poland: (1) descriptive medical diagnoses; (2) discharge recommendations; (3) echocardiography report (if performed). Set of discharge summaries was analyzed with both the conventional (manual) method and the algorithm to demonstrate the process of acquisition of basic characteristics of the cohort of patients with atrial fibrillation/flutter.
Methods: Discharge summaries (validation dataset) of 400 patients hospitalized at one cardiology department were analyzed (1) automatically and (2) manually coded into database by a healthcare professional, utilizing proprietary developed annotation tool to accelerate annotation process, minimize errors and calculate total effective data acquisition time.
Results: The time of manual and automatic data analysis was 13:08 and 0:21 hours, respectively. The overall macroaveraged F1-score for automatic detection with manual detection as a reference was: 0.924 for diagnoses, 0.983 for drug groups and 0.988 for echo parameter retrieval indicating high agreement. Some differences between the 2 classifications were noted, but did not reach statistical significance. There were total of 181 errors, within a total of 9,535 identified parameters (diagnoses, medical substances, or echo parameters) analyzed. Manual qualitative analysis revealed 65.8% of them related to random algorithm errors, 21.5% to manual annotation errors and 12.7% errors related to a lack of advanced context analysis.
Conclusions: The utilization of the algorithm greatly reduced the time required for basic characteristics of the group acquisition without significantly compromising the quality of the data. Automatic detection of retrospective study cohort through application of text processing techniques from electronic health records is promising and feasible. Further progress can be made with utilization of large language models due to superior context awareness. ❑
Figure 1